Publications
Highlights
(For a full list of publications see below or at Google Scholar)
|
Wu G, Zaker A, Ebrahimi A, Tripathi S, Mer AS
|
|
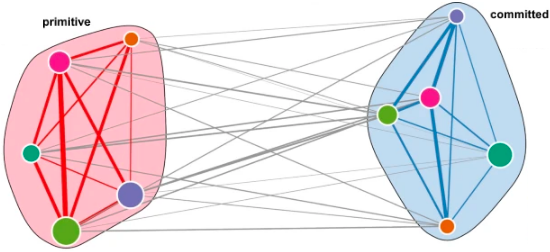
Mer AS, Heath E, Tonekaboni S, Garcia-Prat L, Shlush L, Voisin V, Bader G, Lupien M, Dick J, Minden M, Schimmer A, Haibe-Kains B Nature Communications, Jan. 2021
|
|
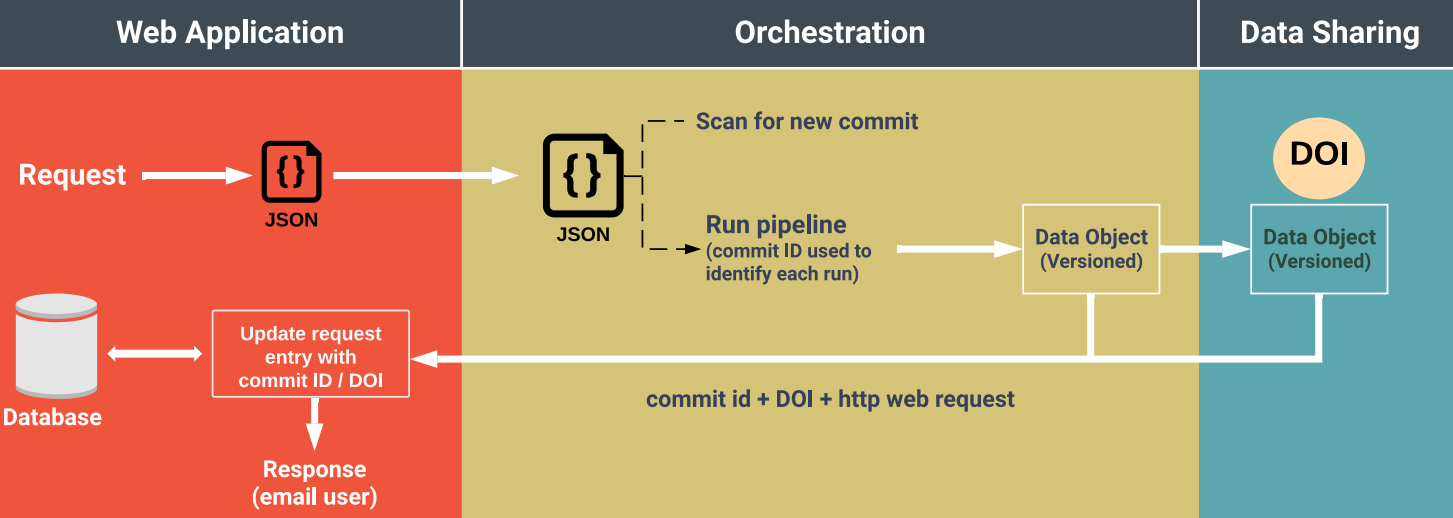
Mammoliti A, Smirnov P, Nakano M, Safikhani Z, Eeles C, Seo H, Nair SK, Mer AS, Ho C, Beri G, Kusko R, MAQC Society, Haibe-Kains B Nature Communications, Oct. 2021
|
|
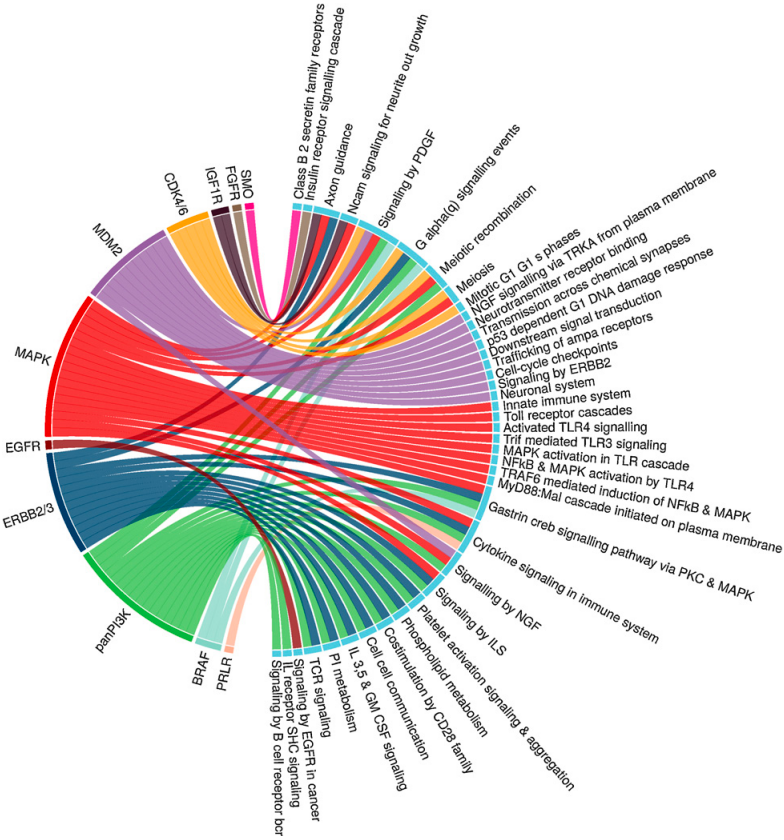
Mer AS, Ba-alawi W, Smirnov P, Wang XY, Brew B, Ortmann J, Tsao MS, Cescon D, Goldenberg A, Haibe-Kains B,
|
Full List of publications
Publications
|
Spotlight on nuclear PD-L1 in ovarian cancer chemoresistance: hidden but mighty Asare-Werehene M, Zaker A, Tripathi S, Communal L, Carmona E, Mes-Masson A-M, Tsang BK and Mer AS Front. Immunol. 16:1543529, 2025 |
|
From Data Annotation to AI Prediction: Streamlining Histopathology Analysis in Acute Respiratory Distress Syndrome Ebrahimi A, Zarei MR, Kuhar E, Karunamurthy P, Pillagawa K, Jahandideh F, Komeili M, Lalu M, Mer AS Proceedings of the Canadian Conference on Artificial Intelligence, 2025 |
|
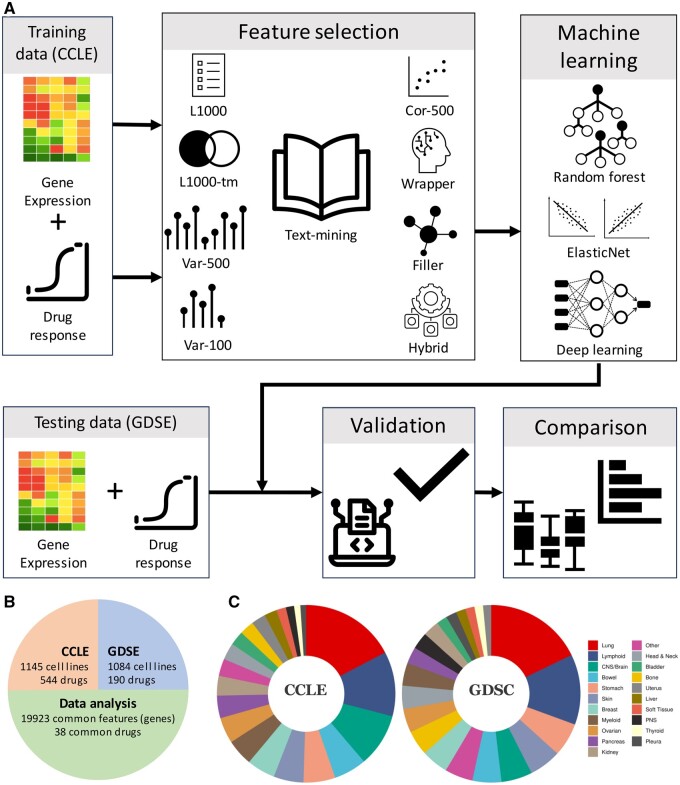
Text‑mining‑based feature selection for anticancer drug response prediction Wu G, Zaker A, Ebrahimi A, Tripathi S, Mer AS Bioinformatics Advances, 2024 |
|
Characterization of the diversity of type IV secretion system‑encoding plasmids in Acinetobacter Nasser F, Gaudreau A, Lubega S, Zaker A, Xia X, Mer AS, D’Costa VM Emerging Microbes & Infections, 2024 |
|
Structural insights into an atypical histone binding mechanism by a PHD finger Grégoire S, Grégoire J, Yang Y, Capitani S, Joshi M, Sarvan S, Zaker A, Ning Z, Figeys D, Ulrich K, Brunzelle JS, Mer AS, Couture JF Structure 2024 |
|
Large‑scale phenogenomic analysis of human cancers uncovers frequent alterations affecting SMC5/6 complex components in breast cancer Roy S, Zaker A, Mer AS(co corresponding author) , D’Amours D NAR Cancer 2023 |
|
Spatiotemporal modeling of chemoresistance evolution in breast tumors uncovers dependencies on SLC38A7 and SLC46A1 Audet‑Delage Y, St‑Louis C, Minarrieta L, McGuirk S, Kurreal I, Annis MG, Mer AS, Siegel PM, St‑Pierre J. Cell reports 2023 |
|
Differential DNA damage repair and PARP inhibitor vulnerability of the mammary epithelial lineages Kim H, Aliar K, Tharmapalan P, McCloskey CW, Kuttanamkuzhi A, Grünwald BT, Palomero L, Mahendralingam MJ, Waas M, Mer AS, Elliott MJ, Zhang B, Al‑Zahrani KN, Langille ER, Parsons M, Narala S, Hofer S, Waterhouse PD, Hakem R, Haibe‑Kains B, Kislinger T, Schramek D, Cescon DW, Pujana MA, Berman HK, Khokha R. Cell reports 2023 |
|
Bimodal gene expression in cancer patients provides interpretable biomarkers for drug sensitivity Ba-Alawi W, Nair SK, Li B., Mammoliti A., Smirnov P, Mer AS, Linda PZ, Haibe-Kains B Cancer Research, 2022 |
|
IPO11 regulates the nuclear import of BZW1/2 and is necessary for AML cells and stem cells Nachmias B, Khan DH, Voisin V, Mer AS, Thomas GE, Segev N, St-Germain J et al. Leukemia Feb. 2022 |
|
PharmacoDB 2.0: improving scalability and transparency of in vitro pharmacogenomics analysis Feizi N, Nair SK, Smirnov P, Beri G, Eeles C, Esfahani PN, Nakano M, Tkachuk D, Mammoliti A, Gorobets E, Mer AS, Lin E, Yu Y, Martin S, Hafner M, Haibe-Kains B Nucleic Acids Res. 2022 Jan 7 |
|
Ontogeny and Vulnerabilities of Drug-Tolerant Persisters in HER2+ Breast Cancer Chang CA, Jen J, Jiang S, Sayad A, Mer AS, Brown KR, Nixon AM, Dhabaria A, Tang KH, Venet D, Sotiriou C, Deng J, Wong KK, Adams S, Meyn P, Heguy A, Skok JA, Tsirigos A, Ueberheide B, Moffat J, Singh A, Haibe-Kains B, Khodadadi-Jamayran A, Neel BG Cancer discovery, Dec. 2021 |
|
Assessing therapy response in patient-derived xenografts Ortmann J, Rampášek L, Tai E, Mer AS, Shi R, Stewart EL, Mascaux C, Fares A, Pham N, Beri G, Eeles C, Tkachuk D, Ho S, Sakashita S, Weiss J, Jiang X, Liu G, Cescon D, O’Brien C, Guo S, Tsao M, Haibe-Kains B, Goldenberg A Science Translational Medicine Nov. 2021 |
|
Novel subtypes of NPM1-mutated AML with distinct outcome Mer AS, Minden M, Haibe-Kains B, Schimmer A Molecular & Cellular Oncology, 2021 |
|
Orchestrating and sharing large multimodal data for transparent and reproducible research Mammoliti A, Smirnov P, Nakano M, Safikhani Z, Eeles C, Seo H, Nair SK, Mer AS, Ho C, Beri G, Kusko R, MAQC Society, Haibe-Kains B Nature Communications, Oct. 2021 |
|
Proteogenomic characterization of pancreatic ductal adenocarcinoma Cao L, Huang C, Cui DZ et al. (including Mer AS) Cell, 2021 |
|
Drug Sensitivity Prediction From Cell Line-Based Pharmacogenomics Data: Guidelines for Developing Machine Learning Models Noghabi HS, Jahangiri ST, Smirnov P, Hon C, Mammoliti A, Nair SK, Mer AS, Ester M, Haibe-Kains B Briefings in Bioinformatics 2021 |
|
Biological and therapeutic implications of a unique subtype of NPM1 mutated AML Mer AS, Heath E, Tonekaboni S, Garcia-Prat L, Shlush L, Voisin V, Bader G, Lupien M, Dick J, Minden M, Schimmer A, Haibe-Kains B Nature Communications, Jan. 2021 |
|
Colorectal Cancer Cells Enter a Diapause-like DTP State to Survive Chemotherapy Rehman S, Haynes J, Collignon E, Brown K, Wang Y, Nixon A, Bruce J, Wintersinger J, Mer AS, Lo E, Leung C, Lima-Fernandes E, Pedley N, Soares F, McGibbon S, He H, Pollet A, Pugh T, Haib-Kains B, Morris Q, Ramalho-Santos M, Goyal S, Moffat J, O’Brien C Cell, 2021 |
|
Integrative Pharmacogenomics Analysis of Patient Derived Xenografts Mer AS, Ba-alawi W, Smirnov P, Wang XY, Brew B, Ortmann J, Tsao MS, Cescon D, Goldenberg A, Haibe-Kains B, Cancer Research, May 2019 |
|
Machine Learning for Biomarker Discovery in Cancer Pharmacogenomics Data Mer AS, Smirnov P, Haibe-Kains B Proceedings of 10th ACM BCB 2019 |
|
Organoid cultures as preclinical models of non-small cell lung cancer Shi R, Radulovich N, Ng C, Liu N, Notsuda H, Cabanero M, Martins-Filho SN, Raghavan V, Li Q, Mer AS, Rosen J, Li M, Wang Y, Tamblyn L, Pham N, Haibe-Kains B, Liu G, Moghal N, Tsao MS Clinical Cancer Research, 2019 |
|
Expression levels of long non-coding RNAs are prognostic for AML outcome Mer AS, Lindberg L, Nilsson C, Klevebring D, Wang M, Grönberg H, Lehmann S, Rantalainen M Journal of Hematology & Oncology 11(1), 2018 |
|
Disruption of the anaphase-promoting complex confers resistance to TTK inhibitors in triple-negative breast cancer Thu KL, Silvester J, Elliott MJ, Ba-Alawi W, Duncan MH, Elia AC, Mer AS, Smirnov P, Safikhani Z, Haibe-Kains B and Mak TW PNAS, 2018 |
|
Integrative cancer pharmacogenomics to establish drug mechanism of action: drug repurposing El-Hachem N, Ba-alawi W, Smith I, Mer AS, Haibe-Kains B Pharmacogenomics, Nov. 2017 |
|
Software for the integration of multi-omics experiments in Bioconductor Ramos M, Schiffer L, Re A, Azhar R, Basunia A, Rodriguez C, Chan T, Chapman P, Davis SR, Gomez-Cabrero D, Culhane AC, Haibe-Kains B, Hansen KD, Kodali H, Louis MS, Mer AS, Riester M, Morgan M, Carey V, Waldron L Cancer Research Nov., 2017 |
|
Validation of risk stratification models in acute myeloid leukemia using sequencing-based molecular profiling Wang M, Lindberg J, Klevebring D, Nilsson C, Mer AS, Rantalainen M, Lehmann S, Grönberg H Leukemia, Feb. 2017 |
|
Study design requirements for RNA sequencing-based breast cancer diagnostics Mer AS, Klevebring D, Gronberg H, Rantalainen M Scientific Reports, 2016 |
|
CellWhere: graphical display of interactome networks organized on subcellular localizations Zhu L, Malatras A, Thorley M, Aghoghogbe I, Mer AS, Duguez S, Butler-Browne G, Voit T, Duddy W Nucleic acids research 2015 |
|
mBISON: Finding miRNA target over-representation in gene lists from ChIP-sequencing data Gebhardt ML, Mer AS, Andrade-Navarro MA BMC Research Notes, April 2015 |
|
A novel approach for protein subcellular location prediction using amino acid exposure Mer AS, Andrade-Navarro MA BMC Bioinformatics. 2013 Nov 28;14(1):342 |
|
MicroRNAs Modulate the Dynamics of the NF-𝜅B Signaling Pathway Vaz C, Mer AS, Bhattacharya A, Ramaswamy R PloS one, 6(11), 2011 |
Book Chapters
|
Applications of Computational Systems Biology in Cancer Signaling Pathways. In Unravelling Cancer Signaling Pathways: A Multidisciplinary Approach Sandhu V, Manem VS, Mer AS, Kure EH, Haibe-Kains B Springer Nature Singapore, 2019 |
|
PRC1-mediated gene silencing in pluripotent ES cells: function and evolution. Epigenetic Mechanisms in Cellular Reprogramming. In Epigenetic Mechanisms in Cellular Reprogramming (pp. 141-166) Becker M, Mah N, Zdzieblo D, Li X, Mer AS, Andrade-Navarro MA, Müller AM Springer Berlin Heidelberg 2015 |
